Authors: Kim van Dongen, Msc
Inge Thijssen-van Loosdregt, PhD
CytoSMART® Technologies B.V., Eindhoven, The Netherlands
Introduction
Transient transfection is a widely used technique for introducing foreign nucleic acids (DNA/RNA) into eukaryotic cells. It is a common method to study and modulate gene expression for a finite period of time, which allows for a better understanding of gene functions and products in cells. Nowadays, there are numerous methods to introduce foreign material into cells, each with its own efficacy.
A particularly useful transfection method is the BacMam virus expression system. This technology uses a baculovirus – an insect virus modified with a mammalian recognizable promotor – to help efficiently deliver and (over)express genes in mammalian cells (Baculovirus – Mammalian cells). Compared with other transfection systems (e.g. plasmid transfection), the BacMam system offers rapid gene delivery, simple workflows and a safe alternative due to its non-replicative nature and lack of cytotoxic effects on mammalian cells [1,2].
The BacMam system has been used for a wide variety of applications such as modification and insertion of genes [3], vaccine development [4], and fluorescent labeling of specific cell structures or cellular markers [5]. Labeling structures in living cells is possible with a commercially available format of BacMam (Invitrogen CellLightsTM). These reagents contain fusions of fluorescent proteins with signal peptides that direct the fluorescent marker to specific cell structures (e.g. actin). The fluorescent fusion protein is introduced into the cell using a simple transfection method (Fig. 1). Briefly, BacMam particles with the fluorescent fusion construct are taken up via endocytosis and the released DNA is translocated to the nucleus for transcription. Following gene expression, fluorescent proteins can be detected within 24 hours of transfection.
A major advantage of the BacMam system is that a wide range of mammalian cells can be transfected, including cell lines [6], primary cells [7] and stem cells [8]. However, like any transfection method, the BacMam method does not transfect each cell type with equal efficiency, therefore, the transfection conditions should be optimized per cell type and BacMam construct.
In this proof-of-concept study, we demonstrate the use of the incubator-compatible CytoSMART® Lux3 FL fluorescence imager to easily optimize transfection conditions using the BacMam system. The human hepatoma HepG2 cell line was transfected with BacMam reagents containing actin-GFP fusion constructs. The virus concentration, incubation time and cell density at transfection were varied to find the optimal transfection conditions. Thereupon, we analyzed the transfection efficiencies using the integrated cloud-based image analysis algorithms.
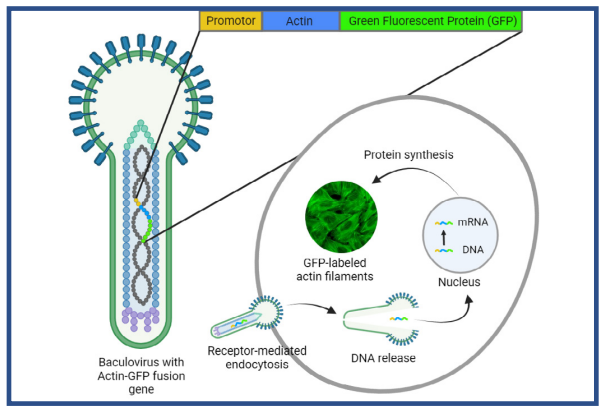
Fig. 1. BacMam-mediated gene delivery. The baculovirus contains DNA of the mammlian promotor and the target protein, coupled to a flouorescent protein (e.g. Green Fluorescent Protein). The BacMam particles enter the cell via endocytosis, after which the DNA is translocated to the nudeus for gene expression. Image modified from [9].
Materials and methods
HepG2 cells were cultured in Advanced DMEM (Gibco) supplemented with 10% FBS (Gibco) and 1% P/S (Gibco). Before transfection, cells were washed with PBS (Gibco), trypsinized and resuspended in phenol red free DMEM (Gibco) supplemented with 10% FBS (Gibco), 2 mM glutamine (Gibco), and 1% P/S (Gibco). Cells were counted using the Corning Cell Counter and transfected with BacMam particles prior to plating. The cells were seeded in a 96-well plate at 16,000 or 24,000 cells/well (100 µL working solution) and incubated with different amounts of BacMam particles/cell (0 (control), 30, 50 or 100).

Fig. 2. Image analysis algorithms. A) Illustration of the confluence algorithm to determine the transfection efficiency, expressed as the confluence ratio of the green fluorescence over the brightfield confluence. B) Illustration of the object count algorithm which annotates and counts fluorescent objects.
Approximately 48 h after seeding, the well plate was placed on the CytoSMART® Lux3 FL inside a cell culture incubator (37°C, 5% CO2). Brightfield and green fluorescence images were taken of each sample2 (two replicates). Images were uploaded to the CytoSMART® Cloud where the transfection efficiency was determined by calculation of the ratio between green fluorescence confluence and brightfield confluence (percentage area covered by cells) using the integrated algorithm (Fig. 2A). Furthermore, the condition with highest transfection efficiency was monitored over time, taking brightfield and green fluorescence images every 30 min for 48 h, and analyzed using the confluence levels and integrated object count feature (Fig. 2B).
Results
Images of the infected cells at 48 hours indicate Actin-GFP fusion protein expression in all transfection conditions (Fig. 3A). The amount of GFP fluorescence was affected by viral concentration at the time of infection, as a higher number of cells express actin GFP upon transfection with the highest BacMam concentration of 100 particles per cell (PPC). Quantification of the transfection efficiency confirmed this observation (Fig. 3B). Interestingly, the highest seeding density (24,000 cells) in combination with high viral loads (100 PPC) resulted in an increase in GFP expression, reaching a transfection efficiency of approximately 70% after 48 h (Fig. 3B).
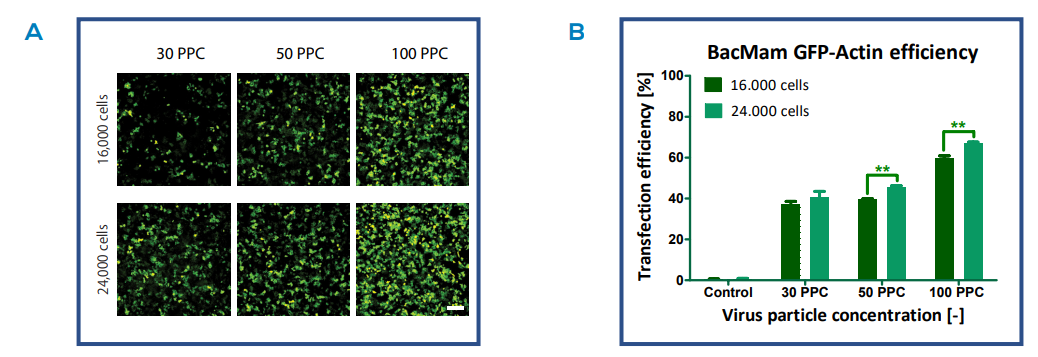
Fig. 3. GFP expression of different transfection conditions. A) Images of HepG2 cells transfected with BacMam Actin virus at different virus particle concentrations (particles per cell (PPC)) and cell densities (16,000 and 24,000 cells/well). The images show the expression of Actin-GFP fusion proteins 48 h after infection. Scale bar indicates 200 µm. B) Quantification of the transfection efficiency 48 h after infection (mean ± sd (n = 2), P ≤ 0.01).
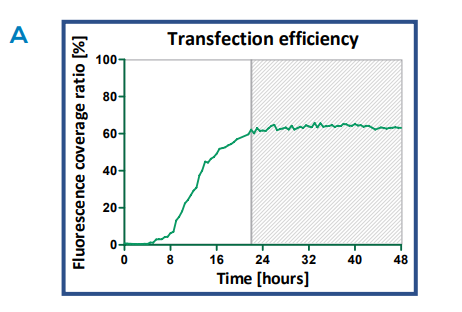

Fig. 4. GFP expression time course. A) The transfection efficiency over time of HepG2 cells transfected with BacMam virus (100 particles per cell and 24,000 cells). The efficiency was determined by calculation of the confluence levels in images taken every 30 minutes over a 48- hour time period. B) Images of the transfected HepG2 cells at 2h time intervals. Upon transfection, Actin-GFP fusion protein is expressed in actin filaments. Scale bar indicates 200 µm.
HepG2 cells transfected using optimal conditions (100 PPC, 24,000 cells) were monitored kinetically over a period of 48 h. As demonstrated in Figure 4, GFP fluorescence can be observed as soon as 6 h (omitting background fluorescence signal). The percentage of fluorescent cells continued to increase up to 22 h, after which the fluorescence level reached a plateau (Fig. 4A) As the transfection progressed, more cells expressed GFP and after 12 h the fluorescence became more concentrated, resulting in brighter fluorescent signals (Fig. 4B).
Fluorescence object counting identified an increasing number of green transfected cells over time (Fig. 5). While the transfection efficiency reached a maximum plateau around 22 h (Fig. 4A), the object count continued to increase over time and reached a more constant phase after 33 h. This indicates that the cells continued to proliferate (as confirmed by an increase in brightfield and green confluence (data not shown)), transferring the Actin-GFP DNA into daughter cells, resulting in more fluorescent objects.
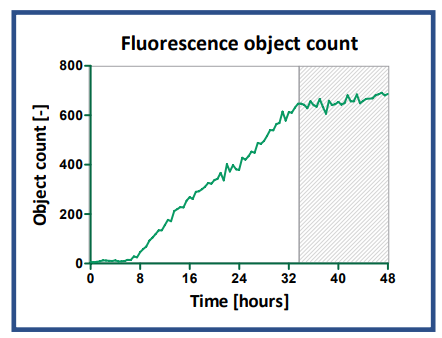
Fig. 6. Fluorescence object count time course. The object number was determined with the integrated object count feature in the Cloud, detecting green fluorescent signals (Actin-GFP) in images taken every 30 minutes over a 48-hour time period.
Discussion
With the BacMam Technology, one can easily label cellular structures with a fluorophore, whose fluorescence expression can be analyzed with a fluorescence imager and optimized if needed. Optimization of the transfection conditions is necessary to ensure a high transfection efficiency with limited cytotoxicity. In this study, we used the CytoSMART® Lux3 FL to optimize the conditions of BacMam Actin-GFP transfection of HepG2 cells. The experiments showed that the imaging system can easily and quickly examine multiple transfection conditions at fixed time points by taking a snapshot of each condition. The confluence algorithm in the CytoSMART® Cloud was then used to calculate the transfection efficiency. Although only one snapshot was analyzed in time, we were able to find the optimal cell density (24,000 cells/well) and virus concentration (100 PPC). In comparison, the manufacturer recommends 10-50 PPC, which is lower than we used.
In addition, kinetic measurements were performed to provide data about the transfection process in real-time. The CytoSMART® Lux3 FL allowed live-cell imaging of our transfected cells inside the incubator, ensuring a constant and optimal culture environment. Confluence levels of the brightfield and green fluorescence channel were automatically determined in the Cloud and used to calculate transfection efficiency. The temporal increase in fluorescence showed that the maximal transfection efficiency was achieved at 22 h, which is consistent with literature [10]. The number of fluorescent objects kept increasing after the transfection efficiency reached a plateau, indicating that the transfected cells were able to proliferate. Therefore, we can assume that the concentration of 100 PPC is not cytotoxic to HepG2 cells.
Conclusion
In this study, we have shown a proof-of-concept for the use of fluorescence live-cell imaging in optimizing transfection conditions of the BacMam system. The expression of BacMam constructs – specifically targeting actin-GFP – could be easily monitored with the CytoSMART® Lux3 FL. Next to this, the integrated confluence measurements and fluorescent object counting provide two different methods to easily determine the transfection efficiency.
References
[1] Fung, K. L., Kapoor, K., Pixley, J. N., Talbert, D. J., Kwit, A. D., Ambudkar, S. V., & Gottesman, M. M. (2016). Using the BacMam baculovirus system to study expression and function of recombinant efflux drug transporters in polarized epithelial cell monolayers. Drug Metabolism and Disposition, 44(2), 180-188.
[2] Lee, H. P., & Hu, Y. C. (2007). Expression in mammalian cells using BacMam viruses. Expression Systems: Methods Express. Scion Publishing Limited: Plymouth, UK.
[3] Hindriksen, S., Bramer, A. J., Truong, M. A., Vromans, M. J., Post, J. B., Verlaan-Klink, I., ... & Hadders, M. A. (2017). Baculoviral delivery of CRISPR/ Cas9 facilitates efficient genome editing in human cells. PloS one, 12(6), e0179514.
[4] Abe, T., & Matsuura, Y. (2010). Host innate immune responses induced by baculovirus in mammals. Current gene therapy, 10(3), 226-231.
[5] Mansouri, M., Bellon-Echeverria, I., Rizk, A., Ehsaei, Z., Cosentino, C. C., Silva, C. S., ... & Berger, P. (2016). Highly efficient baculovirus-mediated multigene delivery in primary cells. Nature communications, 7(1), 1-13.
[6] Hassan, N. J., Pountney, D. J., Ellis, C., & Mossakowska, D. E. (2006). BacMam recombinant baculovirus in transporter expression: a study of BCRP and OATP1B1. Protein expression and purification, 47(2), 591–598. https://doi.org/10.1016/j.pep.2005.12.008
[7] Grassi, G., Köhn, H., Dapas, B., Farra, R., Platz, J., Engel, S., Cjsareck, S., Kandolf, R., Teutsch, C., Klima, R., Triolo, G., & Kuhn, A. (2006). Comparison between recombinant baculo- and adenoviral-vectors as transfer system in cardiovascular cells. Archives of virology, 151(2), 255–271.
[8] Ho, Y. C., Chung, Y. C., Hwang, S. M., Wang, K. C., & Hu, Y. C. (2005). Transgene expression and differentiation of baculovirus-transduced human mesenchymal stem cells. The journal of gene medicine, 7(7), 860–868.
[9] ThermoFisher Scientific (2021, December). BacMam Technology Overview. https://www.thermofisher.com/nl/en/home/industrial/pharmabiopharma/drug-discovery-development/target-and-lead-identification-and-validation/pathway-biology/cellular-pathway-analysis/bacmamsystem/bacmam-technology-overview.html
[10] Matilainen, H., Rinne, J., Gilbert, L., Marjomäki, V., Reunanen, H., & Oker-Blom, C. (2005). Baculovirus Entry into Human Hepatoma Cells. Journal of Virology, 79(24), 15452–15459.
