Introduction
Cell toxicity, viability and growth are measures widely-used in the fields of cancer research, drug development, and tissue engineering. Monitoring cell growth is commonly done manually, by quickly investigating the confluency of the cells using a bright-field microscope. By using a bright-field microscope with an automated stage, images can be taken at set positions and time intervals which enables quantitative confluency measurements. Another option to measure viability is to measure cell metabolic activity by using e.g. a MTT[1] or Alamar Blue assay [2].
Even though several methods are available to investigate cell toxicity, viability and growth, none one of them is optimal. Manual investigation of confluency can only be used to monitor cells during culture and not when performing experiments since it is labor intensive and not quantitative. Automated microscopes can be used to quantify the confluency, however, this can usually only be done at one position per sample and an incubation box needs to be build around the microscope to ensure the correct culture conditions (temperature and CO2 -level). Metabolic activity assays are quite accurate and easy to perform, however, these assays are generally end-point measurements. Thus, in case measurements over time are necessary, multiple samples are needed which increases the workload and costs of the experiment.
A solution for the limitations of the aforementioned methods would be to image an entire sample at multiple time-points and at optimum culture conditions. The CytoSMART® Omni is an automated live-cell imager that can be placed inside a regular CO2 -incubator for imaging complete well plates over time. This imager allows for monitoring cell culture at optimal conditions, since it is situated inside an incubator. Next to this, the cells are undisturbed because the microscope itself moves at predefined intervals instead of the stage. Furthermore, it scans the entire surface of a culture plate, enabling confluency measurements of the entire sample instead of measurements taken at only one position per sample. Additionally, the (cloud-based) software of the CytoSMART® Omni provides direct storage and confluency analysis of the images.
The aim of this study was to investigate the feasibility of using the CytoSMART® Omni to measure growth of C6 (rat glial tumor) cells seeded at different cell densities.
Materials and Methods
C6 cells were seeded in a 96-well plate at different densities ranging from 1,300 to 84,000 cells/cm2 in DMEM (Invitrogen) supplemented with 10% FBS and 1% penicillin-streptomycin (n=12 for each group). Immediately after seeding, the 96-well plate containing the samples was placed on the CytoSMART® Omni which was placed inside an incubator (37°C and 5% CO2 ). For the duration of 44 hours, the complete well plate was imaged every 4 hours (Fig. 1A). The CytoSMART® image analysis software was used for well detection (Fig. 1B) and determining the confluency of each individual well (Fig. 1C).
Results
The CytoSMART® software was able to detect the confluency of each individual well over time (Fig. 1D and E). Up to a seeding density of 5,300 cells/cm2 , the confluency slightly increased over time. Starting from 10,600 cells/cm2 the cell density gradually increased over time till a maximum confluency of 60%. The two highest seeding densities (42,000 and 84,000 cells/cm2 ) resulted in a rapid increase in confluency in the first 8 hours after which it slightly increased over time. The variation in confluency increased with increasing seeding density.
Discussion
Few methods are available to quantitatively measure cell viability or growth over time. However, the major drawbacks of these methods are that they are either inaccurate (manual inspection of viability), suboptimal (incubator box around automated microscope) or toxic (metabolic activity assays). Therefore, we aimed to investigate the possibility of following cell growth over time using confluency measurements of an entire sample obtained with the CytoSMART® Omni.
In this study we have shown the feasibility of measuring cell growth over time. Depending on the initial seeding density, the confluency increased over time following different slopes. Using a seeding density of 21,000 cells/cm2 resulted in the most steady growth over time, while samples seeded at lower cell densities hardly increased in confluency, and samples seeded at higher cell densities reached a confluency plateau after the first 8 hours of culture.
Using the CytoSMART® Omni it is possible to visualize the whole well plate which leads to analysis of each complete well, instead of just one image per well, as is the case in most conventional microscopes. This results in more accurate data of the cell growth since all cells are analyzed instead of only a part of the cells. Next to this, since the cells did not have to be taken out of the incubator for imaging, the cells did not experience a temperature shock and were thus cultured at the optimum culture conditions. Furthermore, because the same samples are followed over time the data is more accurate compared to performing multiple end-point measurements at different time-points.
Conclusion
Summarizing, in this study, we showed the feasibility of following cell growth using real-time confluency measurements taken with the CytoSMART® Omni. Compared to traditional bright-field live-cell imaging, the CytoSMART® Omni provided the benefit of culturing cells in optimal culture conditions while monitoring confluency changes.
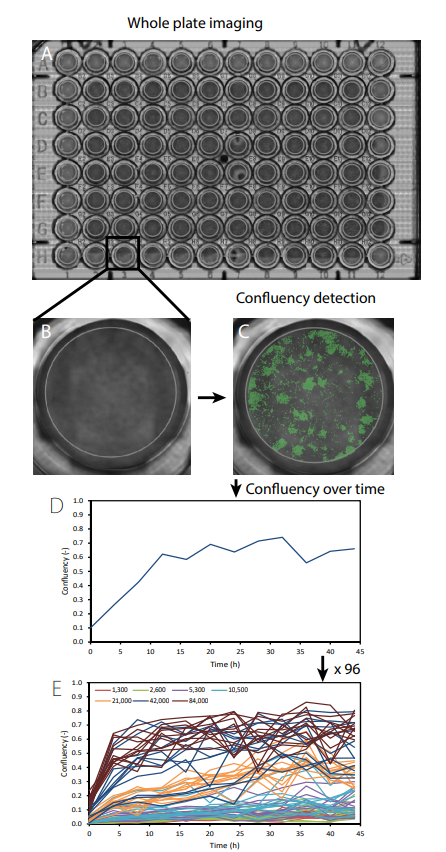
Fig 1. A scan of the whole well plate was made using the CytoSMART® Omni (A). Thereafter individual wells were selected (B) and the confluency was measured (C) over time (D). This was repeated for each well (E). The different colors in (e) represent different seeding densities (cells/cm2).
References
[1] T. Mosmann, “Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays,” J. Immunol. Methods, vol. 65, no. 1–2, pp. 55–63, 1983.
[2] S. Ansar Ahmed, R. M. Gogal, and J. E. Walsh, “A new rapid and simple non-radioactive assay to monitor and determine the proliferation of lymphocytes: an alternative to [3H]thymidine incorporation assay,” J. Immunol. Methods, vol. 170, no. 2, pp. 211–224, 1994.
