Organoids
The term organoids defines three-dimensional (3D) cell structures that contain a multitude of organ-specific cells, that are formed by self-organization and differentiation of stem cells [1], [2]. Organoids are better at representing cellular environments found in vivo than conventional two-dimensional (2D) cell cultures. The ability to self-organize helps organoids mimic the general tissue structure and developmental trajectory found in vivo, and this can often support the differentiation of cell types that would have otherwise been difficult to maintain in 2D culture [3], [4].
Research using organoids gained momentum in 2009 when single stem cells were first used to produce self-organizing intestinal crypt-villus units [5]. A decade later methods are now available that can produce organoids for a variety of organs. Organoids have been produced for the liver [6][7], colon [8][9], intestine [5][10], kidney [11][12], lung [13], prostate [14], pancreas [15], stomach [16], uterus [17], breast tissue [4], thyroid [18], hippocampus [19], cerebral cortex [20], and retina [21].
-------------------------------------
With the CytoSMART Organoid Counting algorithm, researchers can examine organoids of varying shapes and sizes using brightfield image analysis. Learn more about it here.
-------------------------------------
Producing organoids in the lab is dependent on the goals of the researcher, and several considerations will be discussed below.
Organoid culture
Organoid formation and maturation are preceded by a single cell or small cell-cluster expansion and reorganization [2]. There are two main types of organoids based upon the choice of stem cells. The first is derived from PSCs that include both embryonic stem cells (ESCs) and iPSCs and the second type is derived from organ-specific adult stem cells (ASCs) [22]. A variety of workflows have been developed to generate organoids; however, specialized organoid types require unique culture methods, and not all general workflows are appropriate. The choices of cell culture conditions and the 3D matrix are critical for this complex organization. See Figure 1 for an overview of stem cell sources that can be used to generate a multitude of organoids.
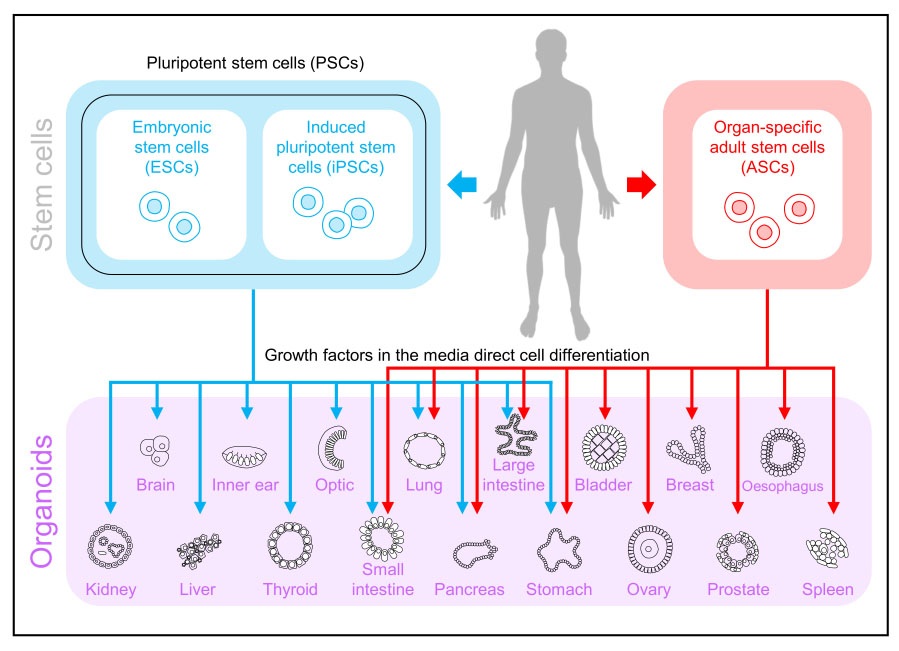
Organoids can be generated by both unguided and guided methods. The unguided approach depends on PSC aggregates to undergo spontaneous morphogenesis and their capacity for intrinsic differentiation, whereas the guided approach induces PSCs to differentiate towards desired lineages using the supplementation of external patterning factors.
The choice between the guided or unguided approach depends on the specific focus of the investigation and the trade-off between diversity and consistency. Both methods can be applied at different stages of organoid development [3].
Growth factors are an essential component of organoid media as these direct the differentiation of the stem cells, through a serial manipulation of combinations of signaling pathways that can generate and sustain specific organoid types [22], [23] (Table 1). For a majority of organoid types, the growth factor requirements include the requirement for Wnt, R-spondin-1 (Rspo), or a combination of both. For example with intestinal stem cells, Rspo and BMP signaling antagonists such as Noggin or Gremlin 1 are needed [24].
Table 1: Growth factors and small molecule inhibitors used in organoid production [25].
Growth factorsNote
EGFUsed for epithelial tissues and promotes tumor growth
FGF10Important for organ development and promotes migration and tumorigenesis
FGF7Promotes growth, invasion, and migration of tumors
HGFPromotes oncogenesis, tumor angiogenesis and invasion
WntA master regulator in regulation of cell development, proliferation, differentiation, adhesion, and polarity
NogginModulates cellular differentiation, proliferation, and apoptosis
RspoRequired for the self-renewal of stem cells and activates Wnt signaling
GastrinStimulates tumor growth
Prostaglandin E2Promotes angiogenesis
NicotinamideVitamin PP is a nutrient that is required for long-term culture of organoids
Neuregulin 1Involved in mammary development and tumorigenesis
Molecule inhibitors
Y27632Reduces the anoikis of dissociated stem cells
A-83-01Suppresses the proliferation of organoids
SB202190Suppresses the proliferation and migration of cancer cells
Organoids are cultured in environments that allow for 3D expansion and self-organization of the cells within the organoid. The most common extracellular matrix (ECM) used is Engelbreth-Hold-Swarm, also referred to as Matrigel, geltrex, and cultrex BME [26]. In certain instances organoids can be cultured successfully in the absence of ECM, such as mammary cell lines that can recapitulate growth and differentiation of a benign mammary tumor [27].
In addition to the ECM, synthetic biomaterials can be engineered to direct the formation of more complex organoids [3]. Proper selection of the matrix used during the culturing process is key in organoid generation. Many more could be selected based on the researcher’s goals, but will not be discussed here. Kratochvil et al. 2019 [26] provide a comprehensive overview of ECM in their review paper.
More complex organoids have been created to improve modeling of inter-regional interactions. An example has been the formation of brain organoids with multiple distinct region identities. This has been achieved by first differentiating human PSCs into different brain region-specific organoids separately, and then fusing them together in a controlled manner to form ‘assembloids’ [3].
Before experiments can be considered, organoids produced in the lab must undergo analysis to confirm that they have the correct tissue structure, and have differentiated correctly.
How are organoids characterized?
Several conventional strategies can be applied to characterize organoids. These techniques include high-resolution microscopy, histology, immunofluorescence, and bulk gene expression assays [28].
Initial characterization can be made using a light microscope. Cell counting is a necessary step for in vitro work involving cell culture. Using the correct organoid concentration will ensure that downstream experiments are reproducible and accurate. Organoid counts are important for monitoring overall health and proliferation rates, seeding for experiments, and preparing for assays [29], [30]. Organoids can be counted using a glass slide or hemocytometer and coverslip [30], [31]. In some instances, organoids can be too large for the hemocytometer and can be counted in droplets on a glass slide [32].
High-resolution microscopy can further be used to assess organoid quantity and morphology. Simple visual observation can determine organoid counts to determine organoid multiplicity before conducting experiments and for experiments that require normalization to controls [23], [33], [34]. Organoid quantity and size can also be determined using the analysis of 8-bit binary images of whole organoid drops [35]. Alternatively, whole-well z-stack images, using a transmitted light inverted microscope with a motorized X/Y scanning stage, can be used to capture all organoids in a well. Organoid culture can then be evaluated based on morphology by taking measurements to quantify variables such as shape and area [36].
Morphological analysis is an important tool, especially when optimizing growth factor concentrations for organoid formation [23]. A variety of techniques exist to assess morphology and differentiation such as brightfield image analysis, and FFPE, and whole-mount staining techniques [36].
Cell viability within organoids can be determined via luminescent cell viability assays [35]. Embedding organoids for histology can be used to observe the interior of samples and ensure that necrosis is not present within the core of an organoid. The differentiation state of the organoid can also be assessed with the use of the sample tissue-specific cell markers and immunofluorescent imaging [36]–[38].
The cell population of an organoid can also be characterized using flow cytometry and bulk gene expression assays. These gene expression assays include qPCR and transcriptome analysis and more recently single-cell RNA sequencing (scRNA-seq) [28], [37], [38].
After the organoids have been characterized they are then ready to be used for downstream applications.
----------------------
For researchers looking for a simplified brightfield microscope that can help in evaluating the success of organoid formation, without having to handle the sample, we recommend the CytoSMART Omni. Create time-lapse videos that can be accessed in real-time. An automated x/y scanner provides a whole-well imaging functionality.
----------------------
What are organoids used for? Applications of organoid technologies
The similarities that organoids share with organs make them a promising research tool for studying more complex biological functions. The use of organoids has already found application in areas such as developmental biology, regenerative medicine, disease modeling, drug discovery, and personalized medicine. The Hubrechts center for Organoid Technology provides a list with possible organoid assays.
Recent advances in organoid research have also improved upon requirements for clinical relevancy. These advances include modeling tumor microenvironments (TME) by co-culturing organoids with immune cells [22], [23], increasing organoid production scale from 106 to 108 cells [41], and high-throughput drug screening methods [42].
In the sections below examples are given for various applications. We also refer to Rossi et al 2018, Kratochvil et al 2019 and Drost, and Clevers 2018, [26], [43], [44] for extended reviews on organoid applications, culturing methods, and cancer models.
Developmental Biology – Tissue Biology
Matured organoids display functional organ sub-structures and have been used to investigate both adult tissue homeostasis and embryonic organ development. Investigation of adult tissue homeostasis using organoids has, for example, provided insight into the structure of intestinal crypts and the nature of the stem-cell niche, that ensures the renewal of the intestine epithelial layer [45]. More recently, a detailed study explored differentiation mapping of the enteroendocrine regulators by using organoids in combination with in vivo models [46].
Studies that focus on embryonic development also benefit from organoid formation. Human induced pluripotent stem cells (iPSCs) can generate organoids that display embryonic morphology and genomic features to draw a parallel to physiological embryonic development [43], [47].
Regenerative medicine
Next to in vitro applications, organoids have been used in transplantation studies to assess regenerative potential. The interest for this is two-fold. Firstly, if transplantation into disease murine models leads to an increased survival rate, organoid functionality is confirmed. Secondly, successful transplantations could open up new avenues for using organoids in regenerative medicine applications as tissue-engineered grafts. In proof-of-concept studies organoids have been used as tissue grafts for both liver [7] and intestine [8]. Building on these insights, efforts were made to establish large-scale production methods of liver buds derived from human PSCs of up to 108 cells [41], and long-term culturing methods for liver organoids [48].
Disease modeling - “Organ Pathology in a Dish”
The therapeutic applications of organoids have been of interest to researchers for several reasons. In disease modeling, organoids generated from human cells can overcome several limitations of animal models [1]. Next to the ethical considerations, some animal models cannot fully recapitulate the human condition, such as with lung tissue [49].
An additional advantage in disease modeling is that organoids provide a platform for modeling pathologies for both embryonic development and adult tissue. Cells containing the pathologies can be generated using gene-editing techniques or be extracted from patients. Pathological conditions in kidney organoids have been generated using a CRISPR/Cas9 knockout system [50]. Cortical organoids, generated from human iPSCs, have been developed to model Miller-Dieker Syndrome [51]. A library of examples of oncogenic organoids is available, many of which are postulated by Drost et al. 2018 [44]. A significantly useful development of organoid culturing for cancer research is the co-culturing of organoids with immune cells [39], [40]. Such methods are important steps towards approaching the physiological TME and provide a platform for immune-based therapies.
Drug discovery and personalized medicine
Organoids are attractive for drug discovery, since they can be produced on a relatively large scale for an array of subtypes capturing disease heterogeneity and stored long term, creating biobanks [52]. Organoid biobanks provide opportunities to test drug safety and efficacy. Toxicology studies can be used to assess safety [53], [54]. For efficacy testing, several high-throughput methods have been developed that can generate results for drug response within a week [42], [55].
When using organoids for personalized medicine, patient-to-patient differences could be captured, but response indications should be producible with clinically relevant timescales. One of the first examples of personalized medicine assays using organoids is that of the forskolin swelling assay, in which drugs for cystic fibrosis can be tested on patient-by-patient efficacy [56]. Other personalized testing platforms using organoids have been developed for kidney [57] and endometrial organoids [17], to give a few examples. Organoids derived from patients have been used to study celiac disease (CD) based on phenotypic dissimilarities and genetic variations between organoids from healthy controls and patients with CD [58].
References
[1] M. A. Lancaster and J. A. Knoblich, “Organogenesis in a dish: Modeling development and disease using organoid technologies,” Science (80-. )., vol. 345, no. 6194, 2014, doi: 10.1126/science.1247125.
[2] Y. Sasai, M. Eiraku, and H. Suga, “In vitro organogenesis in three dimensions: Self-organising stem cells,” Dev., vol. 139, no. 22, pp. 4111–4121, 2012, doi: 10.1242/dev.079590 .
[3] X. Qian, H. Song, and G. Ming, “Brain organoids: advances, applications and challenges,” Development, vol. 146, no. 8, p. dev166074, 2019.
[4] J. M. Rosenbluth et al., “Organoid cultures from normal and cancer-prone human breast tissues preserve complex epithelial lineages,” Nat. Commun., vol. 11, no. 1, pp. 1–14, 2020.
[5] T. Sato et al., “Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche,” Nature, vol. 459, no. 7244, pp. 262–265, 2009, doi: 10.1038/nature07935.
[6] T. Takebe et al., “Vascularized and functional human liver from an iPSC-derived organ bud transplant,” Nature, vol. 499, no. 7459, pp. 481–484, 2013, doi: 10.1038/nature12271.
[7] M. Huch et al., “In vitro expansion of single Lgr5 + liver stem cells induced by Wnt-driven regeneration,” Nature, vol. 494, no. 7436, pp. 247–250, 2013, doi: 10.1038/nature11826.
[8] R. P. Fordham et al., “Transplantation of expanded fetal intestinal progenitors contributes to colon regeneration after injury,” Cell Stem Cell, vol. 13, no. 6, pp. 734–744, 2013, doi: 10.1016/j.stem.2013.09.015.
[9] J. O. Múnera et al., “Differentiation of Human Pluripotent Stem Cells into Colonic Organoids via Transient Activation of BMP Signaling,” Cell Stem Cell, vol. 21, no. 1, pp. 51-64.e6, 2017, doi: 10.1016/j.stem.2017.05.020.
[10] H. Uchida et al., “A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells,” JCI Insight, vol. 2, no. 1, pp. 1–13, 2017, doi: 10.1172/jci.insight.86492.
[11] S. V. Kumar et al., “Kidney micro-organoids in suspension culture as a scalable source of human pluripotent stem cell-derived kidney cells,” Dev., vol. 146, no. 5, 2019, doi: 10.1242/dev.172361.
[12] Z. Li et al., “3D Culture Supports Long-Term Expansion of Mouse and Human Nephrogenic Progenitors,” Cell Stem Cell, vol. 19, no. 4, pp. 516–529, 2016, doi: 10.1016/j.stem.2016.07.016.
[13] B. R. Dye et al., “In vitro generation of human pluripotent stem cell derived lung organoids,” Elife, vol. 2015, no. 4, pp. 1–25, 2015, doi: 10.7554/eLife.05098.
[14] J. Drost et al., “Organoid culture systems for prostate epithelial and cancer tissue,” Nat. Protoc., vol. 11, p. 347, Jan. 2016.
[15] L. Huang et al., “Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids,” Nat. Med., vol. 21, no. 11, pp. 1364–1371, 2015, doi: 10.1038/nm.3973.
[16] T. R. Broda, K. W. McCracken, and J. M. Wells, “Generation of human antral and fundic gastric organoids from pluripotent stem cells,” Nat. Protoc., vol. 14, no. 1, pp. 28–50, 2019, doi: 10.1038/s41596-018-0080-z.
[17] M. Boretto et al., “Patient-derived organoids from endometrial disease capture clinical heterogeneity and are amenable to drug screening,” Nat. Cell Biol., vol. 21, no. 8, pp. 1041–1051, 2019, doi: 10.1038/s41556-019-0360-z.
[18] Y. Saito et al., “Development of a functional thyroid model based on an organoid culture system,” Biochem. Biophys. Res. Commun., vol. 497, no. 2, pp. 783–789, 2018, doi: 10.1016/j.bbrc.2018.02.154.
[19] H. Sakaguchi et al., “Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue,” Nat. Commun., vol. 6, 2015, doi: 10.1038/ncomms9896.
[20] M. A. Lancaster et al., “Cerebral organoids model human brain development and microcephaly,” Nature, vol. 501, no. 7467, pp. 373–379, 2013, doi: 10.1038/nature12517.
[21] S. Kim et al., “Generation, transcriptome profiling, and functional validation of cone-rich human retinal organoids,” Proc. Natl. Acad. Sci. U. S. A., vol. 166, no. 22, pp. 10824–10833, 2019, doi: 10.1073/pnas.1901572116.
[22] J. van der Vaart and H. Clevers, “Airway organoids as models of human disease,” J. Intern. Med., 2020.
[23] M. Urbischek, H. Rannikmae, T. Foets, K. Ravn, M. Hyvönen, and M. de la Roche, “Organoid culture media formulated with growth factors of defined cellular activity,” Sci. Rep., vol. 9, no. 1, pp. 1–11, 2019.
[24] A. Merenda, N. Fenderico, and M. M. Maurice, “Wnt signaling in 3D: Recent advances in the applications of intestinal organoids,” Trends Cell Biol., vol. 30, no. 1, pp. 60–73, 2020.
[25] H. Xu, X. Lyu, M. Yi, W. Zhao, Y. Song, and K. Wu, “Organoid technology and applications in cancer research,” J. Hematol. Oncol., vol. 11, no. 1, p. 116, 2018.
[26] M. J. Kratochvil, A. J. Seymour, T. L. Li, S. P. Paşca, C. J. Kuo, and S. C. Heilshorn, “Engineered materials for organoid systems,” Nat. Rev. Mater., vol. 4, no. 9, pp. 606–622, 2019, doi: 10.1038/s41578-019-0129-9.
[27] S. Florian, Y. Iwamoto, M. Coughlin, R. Weissleder, and T. J. Mitchison, “A human organoid system that self-organizes to recapitulate growth and differentiation of a benign mammary tumor,” Proc. Natl. Acad. Sci., vol. 116, no. 23, pp. 11444–11453, 2019.
[28] H. Wu and B. D. Humphreys, “Single Cell Sequencing and Kidney Organoids Generated from Pluripotent Stem Cells,” Clin. J. Am. Soc. Nephrol., vol. 15, no. 4, pp. 550–556, 2020.
[29] K. Ongena, C. Das, J. L. Smith, S. Gil, and G. Johnston, “Determining cell number during cell culture using the scepter cell counter,” JoVE (Journal Vis. Exp., no. 45, p. e2204, 2010.
[30] T. Grabinger et al., “Ex vivo culture of intestinal crypt organoids as a model system for assessing cell death induction in intestinal epithelial cells and enteropathy,” Cell Death Dis., vol. 5, no. 5, pp. e1228--e1228, 2014.
[31] D. E. Rothschild, T. Srinivasan, L. A. Aponte-Santiago, X. Shen, and I. C. Allen, “The ex vivo culture and pattern recognition receptor stimulation of mouse intestinal organoids,” JoVE (Journal Vis. Exp., no. 111, p. e54033, 2016.
[32] H. E. Francies, A. Barthorpe, A. McLaren-Douglas, W. J. Barendt, and M. J. Garnett, “Drug Sensitivity Assays of Human Cancer Organoid Cultures,” in Organoids: Stem Cells, Structure, and Function, K. Turksen, Ed. New York, NY: Springer New York, 2019, pp. 339–351.
[33] Y. Fujimichi, K. Otsuka, M. Tomita, and T. Iwasaki, “An Efficient Intestinal Organoid System of Direct Sorting to Evaluate Stem Cell Competition in Vitro,” Sci. Rep., vol. 9, no. 1, pp. 1–9, 2019.
[34] C. P. Santos et al., “Urothelial organoids originating from Cd49f high mouse stem cells display Notch-dependent differentiation capacity,” Nat. Commun., vol. 10, no. 1, pp. 1–17, 2019.
[35] K. E. Boonekamp et al., “Long-term expansion and differentiation of adult murine epidermal stem cells in 3D organoid cultures,” Proc. Natl. Acad. Sci., vol. 116, no. 29, pp. 14630–14638, 2019.
[36] T. McCray, Z. Richards, J. Marsili, G. S. Prins, and L. Nonn, “Handling and assessment of human primary prostate organoid culture,” JoVE (Journal Vis. Exp., no. 143, p. e59051, 2019.
[37] M. Dossena et al., “Standardized GMP-compliant scalable production of human pancreas organoids,” Stem Cell Res. Ther., vol. 11, no. 1, pp. 1–12, 2020.
[38] J. Mullenders et al., “Mouse and human urothelial cancer organoids: A tool for bladder cancer research,” Proc. Natl. Acad. Sci., vol. 116, no. 10, pp. 4567–4574, 2019.
[39] J. T. Neal et al., “Organoid Modeling of the Tumor Immune Microenvironment,” Cell, vol. 175, no. 7, pp. 1972-1988.e16, 2018, doi: 10.1016/j.cell.2018.11.021.
[40] K. K. Dijkstra et al., “Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids,” Cell, vol. 174, no. 6, pp. 1586-1598.e12, 2018, doi: 10.1016/j.cell.2018.07.009.
[41] T. Takebe et al., “Massive and Reproducible Production of Liver Buds Entirely from Human Pluripotent Stem Cells,” Cell Rep., vol. 21, no. 10, pp. 2661–2670, 2017, doi: 10.1016/j.celrep.2017.11.005.
[42] N. Phan et al., “A simple high-throughput approach identifies actionable drug sensitivities in patient-derived tumor organoids,” Commun. Biol., vol. 2, no. 1, pp. 1–11, 2019, doi: 10.1038/s42003-019-0305-x.
[43] G. Rossi, A. Manfrin, and M. P. Lutolf, “Progress and potential in organoid research,” Nat. Rev. Genet., vol. 19, no. 11, pp. 671–687, 2018, doi: 10.1038/s41576-018-0051-9.
[44] J. Drost and H. Clevers, “Organoids in cancer research,” Nat. Rev. Cancer, vol. 18, no. 7, pp. 407–418, 2018, doi: 10.1038/s41568-018-0007-6.
[45] T. Sato and H. Clevers, “Growing Self-Organizing Mini-Guts from a Single Intestinal Stem Cell: Mechanism and Applications,” Science (80-. )., vol. 340, no. 6137, pp. 1190–1194, 2013.
[46] H. Gehart et al., “Identification of Enteroendocrine Regulators by Real-Time Single-Cell Differentiation Mapping,” Cell, vol. 176, no. 5, pp. 1158-1173.e16, 2019, doi: 10.1016/j.cell.2018.12.029.
[47] J. Mariani et al., “Modeling human cortical development in vitro using induced pluripotent stem cells,” Proc. Natl. Acad. Sci. U. S. A., vol. 109, no. 31, pp. 12770–12775, 2012, doi: 10.1073/pnas.1202944109.
[48] H. Hu et al., “Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids,” Cell, vol. 175, no. 6, pp. 1591-1606.e19, 2018, doi: 10.1016/j.cell.2018.11.013.
[49] D. Wilkinson et al., “Development of a Three-Dimensional Bioengineering Technology to Generate Lung Tissue for Personalized Disease Modeling,” Stem Cells Transl. Med., vol. 4, pp. 1–11, 2016, doi: 10.5966/sctm.2016-0192.
[50] B. S. Freedman et al., “Modelling kidney disease with CRISPR-mutant kidney organoids derived from human pluripotent epiblast spheroids,” Nat. Commun., vol. 6, no. May, pp. 1–13, 2015, doi: 10.1038/ncomms9715.
[51] V. Iefremova et al., “An Organoid-Based Model of Cortical Development Identifies Non-Cell-Autonomous Defects in Wnt Signaling Contributing to Miller-Dieker Syndrome,” Cell Rep., vol. 19, no. 1, pp. 50–59, 2017, doi: 10.1016/j.celrep.2017.03.047.
[52] N. Sachs et al., “A Living Biobank of Breast Cancer Organoids Captures Disease Heterogeneity,” Cell, vol. 172, no. 1–2, pp. 373-386.e10, 2018, doi: 10.1016/j.cell.2017.11.010.
[53] M. Takasato et al., “Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis,” Nature, vol. 526, p. 564, Oct. 2015.
[54] W. R. Proctor et al., “Utility of spherical human liver microtissues for prediction of clinical drug-induced liver injury,” Arch. Toxicol., vol. 91, no. 8, pp. 2849–2863, 2017, doi: 10.1007/s00204-017-2002-1.
[55] T. Zhou et al., “High-Content Screening in hPSC-Neural Progenitors Identifies Drug Candidates that Inhibit Zika Virus Infection in Fetal-like Organoids and Adult Brain,” Cell Stem Cell, vol. 21, no. 2, pp. 274-283.e5, 2017, doi: 10.1016/j.stem.2017.06.017.
[56] S. F. Boj et al., “Forskolin-induced swelling in intestinal organoids: An in vitro assay for assessing drug response in cystic fibrosis patients,” J. Vis. Exp., vol. 2017, no. 120, pp. 1–12, 2017, doi: 10.3791/55159.
[57] F. Schutgens et al., “Tubuloids derived from human adult kidney and urine for personalized disease modeling,” Nat. Biotechnol., vol. 37, no. 3, pp. 303–313, 2019, doi: 10.1038/s41587-019-0048-8.
[58] W. Dieterich, M. F. Neurath, and Y. Zopf, “Intestinal ex vivo organoid culture reveals altered programmed crypt stem cells in patients with celiac disease,” Sci. Rep., vol. 10, no. 1, pp. 1–10, 2020.
[59] O. Kopper et al., “An organoid platform for ovarian cancer captures intra- and interpatient heterogeneity,” Nat. Med., vol. 25, no. 5, pp. 838–849, 2019, doi: 10.1038/s41591-019-0422-6.
[60] S. F. Roerink et al., “Intra-tumour diversification in colorectal cancer at the single-cell level,” Nature, vol. 556, no. 7702, pp. 437–462, 2018, doi: 10.1038/s41586-018-0024-3.
Related Products
There are currently no products tagged to this resource.